K-Means¶
The code is based upon the Wikipedia description [wikikmeans]. The objective of the algorithm is to minimize the intra-cluster variance, given by

where 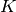 is the total number of clusters,
is the total number of clusters,  the set of points in the
the set of points in the  ‘th cluster, and
‘th cluster, and  the center of the
the center of the  ‘th cluster.
‘th cluster.
When k-means has minimized the intra-cluster variance, it might not have found the global minimum of variance. It is therefore a good idea to run the algorithm several times, and use the clustering result with the best intra-cluster variance. The intra-cluster variance can be obtained from the method find_intra_cluster_variance.
| [wikikmeans] | http://en.wikipedia.org/w/index.php?title=K-means_algorithm&oldid=280702005 |
K-Means Example¶
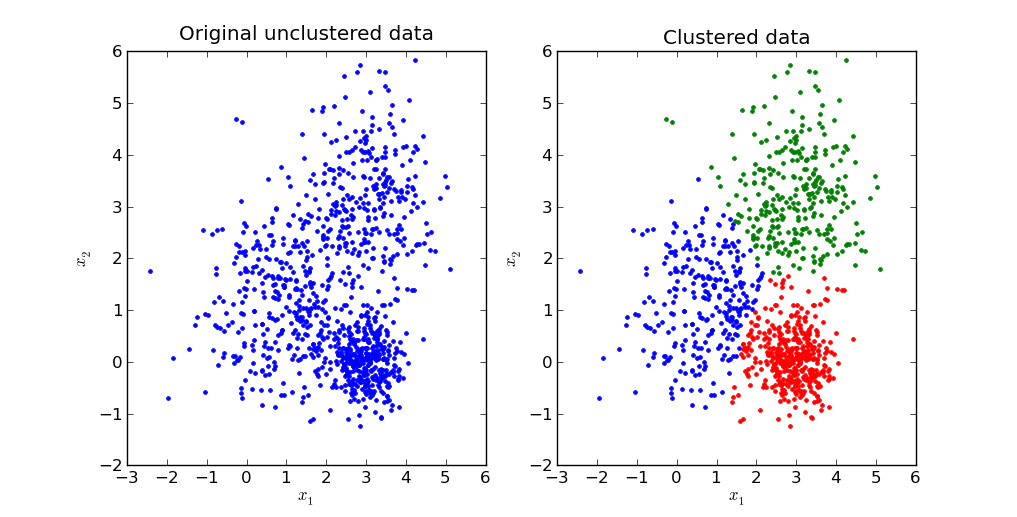
To demostrate the K-means algorithm, we will construct a simple example with three clusters with Gaussian distribution.
from matplotlib.pylab import *
from pypr.clustering.kmeans import *
from pypr.clustering.rnd_gauss_clusters import *
# Make three clusters:
centroids = [ array([1,1]), array([3,3]), array([3,0]) ]
ccov = [ array([[1,0],[0,1]]), array([[1,0],[0,1]]), \
array([[0.2, 0],[0, 0.2]]) ]
X = rnd_gauss_clusters(centroids, ccov, 1000)
figure(figsize=(10,5))
subplot(121)
title('Original unclustered data')
plot(X[:,0], X[:,1], '.')
xlabel('$x_1$'); ylabel('$x_2$')
subplot(122)
title('Clustered data')
m, cc = kmeans(X, 3)
plot(X[m==0, 0], X[m==0, 1], 'r.')
plot(X[m==1, 0], X[m==1, 1], 'b.')
plot(X[m==2, 0], X[m==2, 1], 'g.')
xlabel('$x_1$'); ylabel('$x_2$')
The second plot of this example will give the clustering result from the algorithm.
Application Programming Interface¶
- pypr.clustering.kmeans.find_centroids(X, K, m)¶
Find centroids based on sample cluster assignments.
Parameters : X : KxD np array
K data samples with D dimensionallity
K : int
Number of clusters
m : 1d np array
cluster membership number, starts from zero
Returns : cc : 2d np array
A set of K cluster centroids in an K x D array, where D is the number of dimensions. If a cluster isn’t assigned any points/samples, then it centroid will consist of NaN’s.
- pypr.clustering.kmeans.find_distance(X, c)¶
Returns the euclidean distance for each sample to a cluster center
Parameters : X : 2d np array
Samples are row wise, variables column wise
c : 1d np array or list
Cluster center
Returns : dist : 2d np column array
Distances is returned as a column vector.
- pypr.clustering.kmeans.find_intra_cluster_variance(X, m, cc)¶
Returns the intra-cluster variance.
Parameters : X : 2d np array
Samples are row wise, variables column wise
m : list or 1d np array
Cluster membership number, starts from zero
cc : 2d np array
Cluster centers row-wise, variables column-wise
Returns : dist : float
Intra cluster variance
- pypr.clustering.kmeans.find_membership(X, cc)¶
Finds the closest cluster centroid for each sample, and returns cluster membership number.
Parameters : X : 2d np array
Samples are row wise, variables column wise
cc : 2d np array
Cluster centers row-wise, variables column-wise
Returns : m : 1d array
cluster membership number, starts from zero
- pypr.clustering.kmeans.kmeans(X, K, iter=20, verbose=False, cluster_init='sample', delta_stop=None)¶
Cluster the samples in X using K-means into K clusters. The algorithm stops when no samples change cluster assignment in an itertaion. NOTE: You might need to change the default maximum number of of iterations iter, depending on the number of samples and clusters used.
Parameters : X : KxD np array
K data samples with D dimensionallity.
K : int
Number of clusters.
iter : int, optional
Number of iterations to run the k-means algorithm for.
cluster_init : string, optional
Centroid initialization. The available options are: ‘sample’ and ‘box’. ‘sample’ selects random samples as initial centroids, and ‘box’ selects random values within the space bounded by a box containing all the samples.
delta_stop : float, optional
Use delta_stop to stop the algorithm early. If the change in all variables in all centroids is changed less than delta_stop then the algorithm stops.
verbose : bool, optional
Make it talk back.
Returns : m : 1d np array
cluster membership number, starts from zero.
cc : 2d np array
A set of K cluster centroids in an K x D array, where D is the number of dimensions. If a cluster isn’t assigned any points/samples, then it centroid will consist of NaN’s.